How I stumbled upon a novel genome for a malaria-like parasite of primates
Sometimes in science you come across things that are definitely interesting, and useful, but which you don’t have time to write up properly for one reason or another. I’m going to try to get into the habit of sharing these as blog-posts rather than letting them disappear. Here is one such story.
Update: we published a paper with a fully annotated genome in a big collaboration between the data generators at the University of Oregon and bioinformaticians at the Sanger Institute.
Not long ago I was searching for orthologs of a malaria gene of interest on the NCBI non-redundant database, which allows one to search across the entire (sequenced) tree of life. Here is a recreation of what I saw:

I was surprised to see that nestled among the Plasmodium species was a sequence from a species called Piliocolobus tephrosceles.
A web search revealed this to be the linnaean name for the Ugandan red colobus monkey. Odd that this monkey should have a gene so similar to one from a parasite, eh? But there are some weird misannotations on NCBI. However a bit later I saw the same thing happen again with another gene, and thought something must be going on. I tried more malaria genes and found I could almost always find a strong match in the colobus assembly.
I navigated through NCBI to find some more about this particular monkey.

The page tells us that this monkey from which this genome was sequenced was found here, in the depths of Kibale National Park (it was sequenced for a University of Oregon genome study, in prep.):
It also tells us that the sequence came from whole blood. This methodology seemed to be consistent with researchers serendipitously acquiring sequence from a blood-borne parasite in the process of sequencing the monkey genome.
Just how much of this genome is there in the colobus sample? I downloaded the assembly to find out. The assembly was broken up into 47,644 contigs. My hypothesis was that some of these represented monkey sequence and others represented this sequence of a malaria-like parasite. I calculated the GC content of each contig and plotted a distribution:
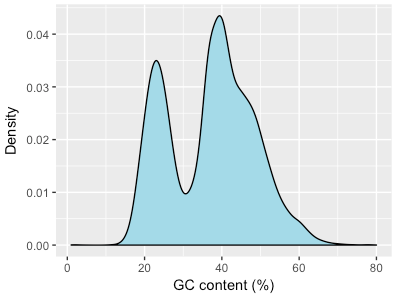
Plasmodium spp. have a much lower GC-content than mammalian species, and from this it looks like whatever species we have here shows this same bias. Just to be sure I also searched each “colobus” contig with Diamond (a faster Blast-like tool) against all protein sequences of the rhesus macaque and of P. vivax. If a contig had a significantly higher score in P. vivax than rhesus it was assigned to that species and vice versa. Overlaying this data was pretty compelling:
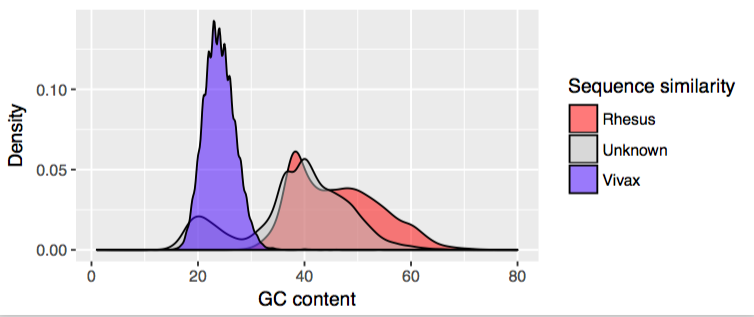
I decided that by taking contigs which were either significantly better matches to P. vivax, or alternatively had a GC content below 24%, I would get a genome which was almost entirely this intriguing species. This gave me 13,460 contigs representing about 30 Mbp of sequence (Plasmodium genomes are 20-30 Mbp so this could potentially be a genome’s worth – although I don’t always get BLAST matches, so I suspect it is somewhat less). I also got in touch with the person named in the NCBI accession to see if they were already investigating these:
hey @theosanderson not specifically on the hepatocystis seqs in the assembly, though collaborators in our group have done some interesting work on hepatocystis in red colobus themselves https://t.co/yLs6ycTYrv
— Noah Simons (@noahdsimons) April 5, 2018
My home institute has released a rather neat web tool called Companion, which allows you to upload a series of genomic contigs and have them annotated with reference to a known genome using a whole pipeline of analysis. (Unfortunately it only accepts up to 3,000 sequences. But you can download it as a Docker container to circumvent that restriction).
Here are the results from its run on these sequences. Companion identifies >2000 genes, including such favourites as circumsporozoite protein, GAP50, PTEX88 and the SERAs. It nicely annotates a pseudochromosome-based version of the genome (although I’m not sure it’s taking full advantage of the synteny at the moment).
So what is this mysterious species?
It clearly isn’t an already sequenced Plasmodium species. A bit of googling suggested to me that it might be some species of Hepatocystis, which are known to infect red Colobus. Hepatocystis seems in some ways to be an unnecessary genus, since the latest phylogenies show that evolutionarily it lies nested within Plasmodium, being more closely related to mammalian Plasmodium species than is the avian malaria parasite Plasmodium gallinaceum. But there are reportedly important differences of approach between the groups – hepatocystis seems to lack the asexual cycle, with replication occurring only in the liver. One might hypothesise that this would lead to substantial loss of intra-erythrocytic genes. Also these beasts are transmitted by midges rather than mosquitoes (that data the result of a decade–long search). There are no other genome-scale sequencing datasets available for hepatocystis.
I searched GenBank for targeted hepatocystis sequences, and found a few, including the caseinolytic protease C gene which has been sequenced from numerous bat isolates. I found the matching sequences for these in the Colobus assembly and then Blasted those sequences back against GenBank to recover a set of sequences to analyse. Constructing a tree with Phylogeny.fr seemed to demonstrate clearly that this is some species of hepatocystis:

The closest relative found here is from a Cynopterus brachyotis bat from Singapore – bats seem to pick up everything!
This is about as far as I’ve got with this. Some thoughts at this point:
- If you are interested in what a specific gene looks like in hepatocystis it is likely that you can find at least part of it in these colobus contigs to compare to. It is very easy to run a BLAST search at NCBI to check.
- Early data-release is awesome! Parasitologists will really benefit from the University of Oregon primate researchers making this data available.
- Someone should really see if we can get a really decent genome from these short read sequences. The library was prepared with Chromium technology and assembled with SuperNova, a 10X assembler. Is it possible these methods are optimised for diploid sequences and that assembling the raw data might enable a better parasite assembly? Or maybe the current assembly is pretty good – apart from the gaps – and missing _Plasmodium _genes are truly missing due to the differences in the hepatocystis lifecycle?
- The depositors of the assembly might wish to partition it into its monkey and hepatocystis portions as I have above, lest a lot of parasite researchers are going to get confused when they find their parasite to be so closely related to a monkey 🙂 Happy to share my assignments!