Abstract
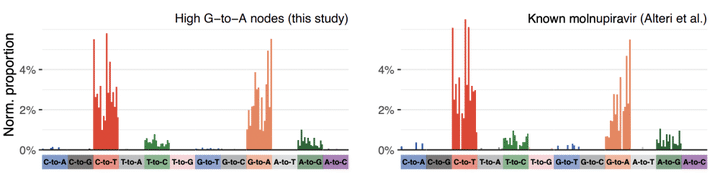
Molnupiravir, an antiviral medication that has been widely used against SARS-CoV-2, acts by inducing mutations in the virus genome during replication. Most random mutations are likely to be deleterious to the virus, and many will be lethal. Molnupiravir-induced elevated mutation rates have been shown to decrease viral load in animal models. However, it is possible that some patients treated with molnupiravir might not fully clear SARS-CoV-2 infections, with the potential for onward transmission of molnupiravir-mutated viruses. We set out to systematically investigate global sequencing databases for a signature of molnupiravir mutagenesis. We find that a specific class of long phylogenetic branches appear almost exclusively in sequences from 2022, after the introduction of molnupiravir treatment, and in countries and age-groups with widespread usage of the drug. We calculate a mutational spectrum from the AGILE placebo-controlled clinical trial of molnupiravir and show that its signature, with elevated G-to-A and C-to-T rates, largely corresponds to the mutational spectrum seen in these long branches. Our data suggest a signature of molnupiravir mutagenesis can be seen in global sequencing databases, in some cases with onwards transmission.
Summary
We analysed global sequence databases to conclusively show that an antiviral drug called molnupiravir has resulted in viable SARS-CoV-2 viruses with significant numbers of mutations, in some cases with onwards transmission of mutated viruses.