Biography
I am an Assistant Professor at the London School of Hygiene and Tropical Medicine where I lead a research group working on large-scale viral genomics.
- Viral Genomics
- Phylogenetics
- Malaria
- Open Source Hardware
- Machine Learning
-
PhD in Malaria Genetics, 2016
Wellcome Sanger Institute
-
BA Natural Sciences, 2011
University of Cambridge
Selected Publications
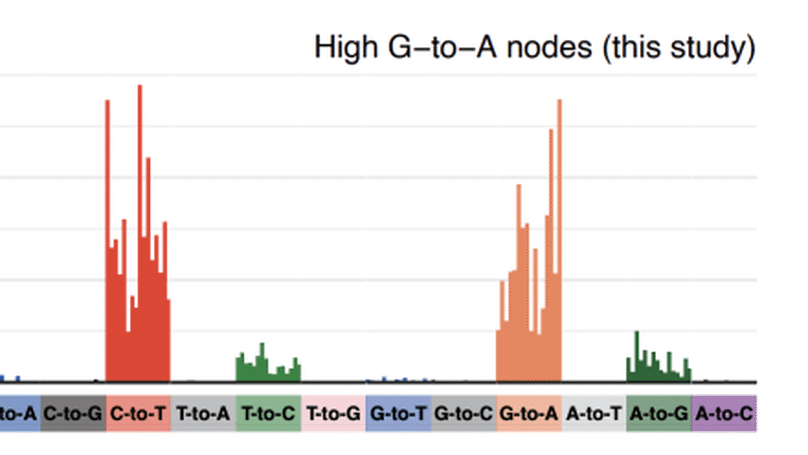
We analysed global sequence databases to conclusively show that an antiviral drug called molnupiravir has resulted in viable SARS-CoV-2 viruses with significant numbers of mutations, in some cases with onwards transmission of mutated viruses.
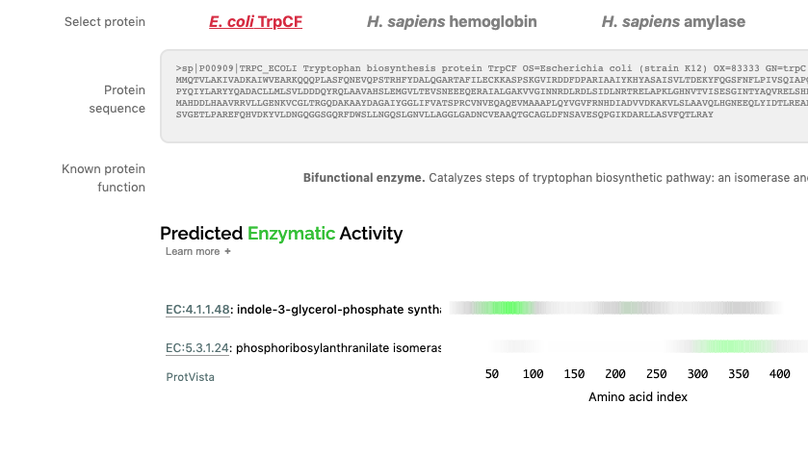
This work, completed as part of my residency at Google AI, uses deep residual networks to predict protein function from amino acid sequences. We show that these networks are able to perform this task effectively, in a way that complements BLAST-based approaches, and that they learn to place protein sequences into a generalised embedding space that facilitates downstream applications. Using TensorFlow JS, we built a tool that performs protein functional inference in the browser, client-side. The paper is presented in an interactive form that allows the reader to explore our work and try the models.
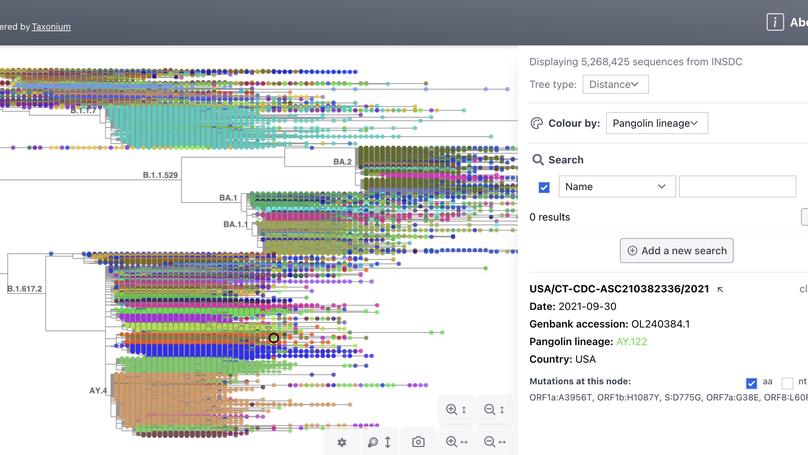
Taxonium is the first tool to allow trees of millions of nodes to be readily explored in the browser.
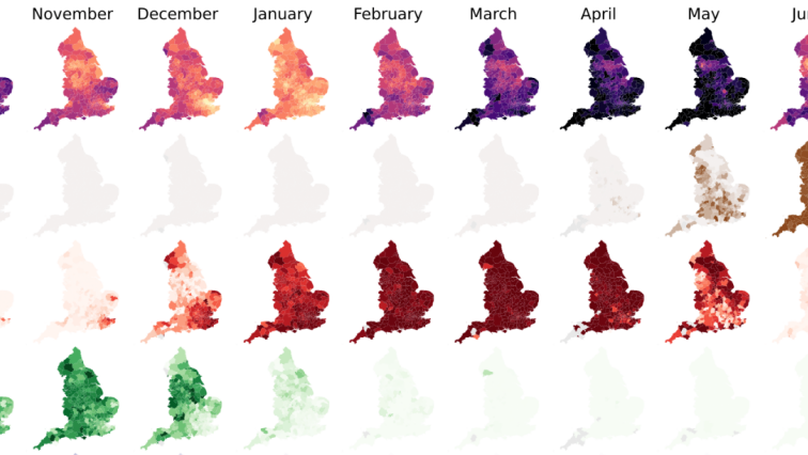
Here we constructed a model of SARS-CoV-2 genomic epidemiology in the UK during 2020-21, chronicling the rise of first the Alpha lineage and the Delta lineage, using data generated at the Sanger Insitute from the sequencing of positive Pillar 2 tests.
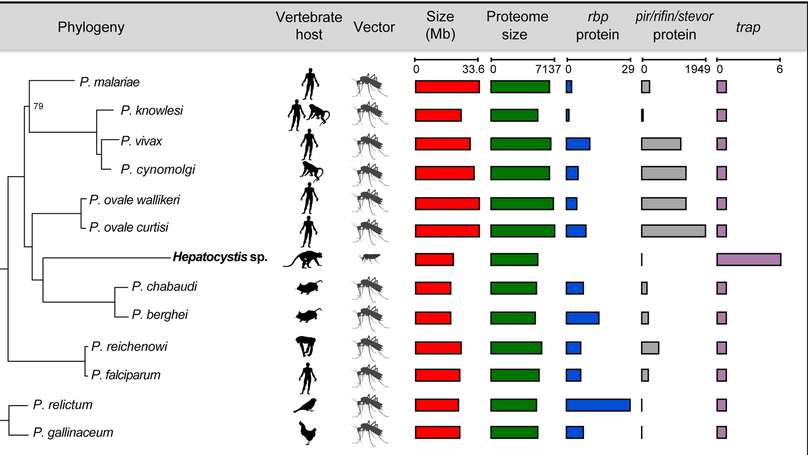
This work began when I did a BLAST search for a malaria parasite gene, and saw a closely matching gene that claimed to be from a monkey. When I investigated further I found that this “monkey genome” contained substantial contamination from a genus of parasite called Hepatocystis that had been lurking in the monkey’s blood. The identification of the first substantial genomic data from this genus, which I initially described in a blog post, triggered a collaborative project between the originators of the data, former colleagues at the Sanger Institute, and myself to characterise this genome revealing the genomic basis of this parasite’s unique biology.
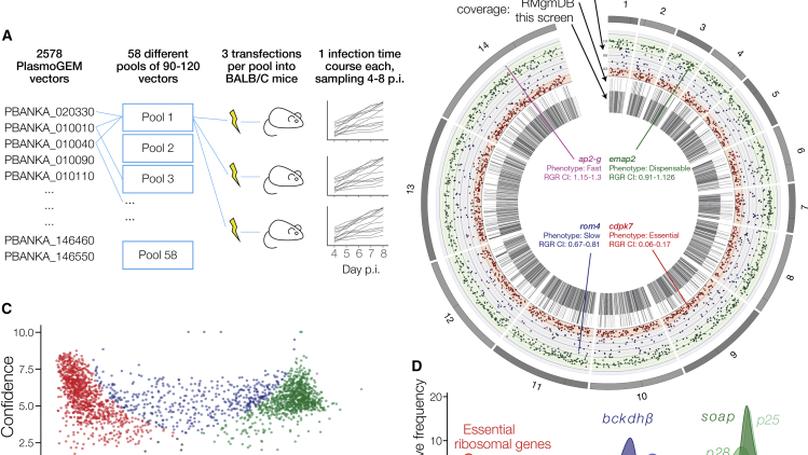
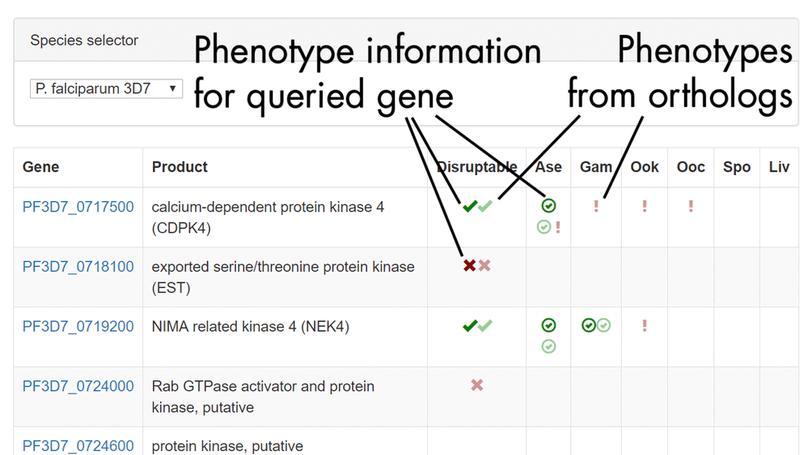
In this work we conducted the first genome-scale genetic screen in a malaria parasite. We found that malaria parasites have require a higher proportion of their genome for normal growth compared to any other eukaryote previously screened. I led the analysis portion of this work, including building the dashboard used by the community to access our phenotype data.
Recent Publications
Recent Posts
Positions held
Contact
- theo@theo.io
- London School of Hygiene and Tropical Medicine, London, WC1E 7HT